Genome size in species of the South American genus Chrysolaena (Vernonieae: Asteraceae) and its relationship with cytogenetic and environmental characteristics
DOI:
https://doi.org/10.30972/bon.3519126Keywords:
Chromosome number, DNA content, 2C value, climatic variables, soil variablesAbstract
Genome size is a species level that often allows taxon delimitation, influences
genetic diversity, and can be related to morphological, ecological, and distribution characteristics.
Chrysolaena H. Rob. (Vernonieae, Asteraceae) is a South American genus, highly cytogenetically
diverse, exhibiting large interspecific and intraspecific chromosomal variation. In this work, the
genome size (2C value) of Chrysolaena species was determined to assess variation among
the different chromosome numbers present in the species, to examine the relationship between
genome size and chromosome numbers, latitude, and environmental variables, and expand
the database for Asteraceae. Linear regression results showed that the 2C value is related to
chromosome number. However, the correlation with latitude and environmental variables is very
low. Evolutionary trends in genome size include both increases and decreases, and polyploidy
is believed to be the main mechanism generating increases in genomic DNA content. The
results of this study constitute an important contribution to the knowledge of genome size in a
representative genus of the Asteraceae family in South America.
Downloads
References
Andréfouët, S., Costello, M. J., Faith, D. P., Ferrier, S., Geller, G. N. & Höft, R. (2008). The GEO Biodiversity Observation Network Concept Document. Geneva.
Angulo, M. B. & Dematteis, M. (2013). Nuclear DNA content in some species of Lessingianthus (Vernonieae, Asteraceae) by flow cytometry. Journal of Plant Research 126: 461-468. https://doi.org/10.1007/s10265-012-0539-x
Ash, N., Jürgens, N., Leadley, P., Alkemade, R., Araújo, M. B. & Asner, G. P. (2009). bioDISCOVERY: Assessing, Monitoring and Predicting Biodiversity.
Baack, E. J., Whitney, K. D. & Rieseberg, L. H. (2005). Hybridization and genome size evolution: timing and magnitude of nuclear DNA content increases in Helianthus homoploid hybrid species. New Phytologist 167: 623-630. https://doi.org/10.1111/j.1469-8137.2005.01433.x
Bancheva, S. & Greilhuber, J. (2006). Genome size in Bulgarian Centaurea s.l. (Asteraceae). Plant Systematics and Evolution 257: 95-117. https://doi.org/10.1007/s00606-005-0384-7
Bennett, M. D. (1972). Nuclear DNA content and minimum generation time in herbaceous plants. Proceedings of the Royal Society B: Biological Sciences 181: 109-135. https://doi.org/10.1098/rspb.1972.0042
Bennett, M. D., Bhandol, P. & Leitch, I. J. (2000). Nuclear DNA amounts in angiosperms and their modern uses-807 new estimates. Annals of Botany 86: 859-909. https://doi.org/10.1006/anbo.2000.1253
Bennett, M. D. & Smith, J. B. (1976). Nuclear DNA amounts in angiosperms. Philosophical Transactions of the Royal Society of London B: Biological Sciences 274: 227-274. https://doi.org/10.1098/rstb.1976.0044
Bennett, M. D. & Leitch, I. J. (2005). Nuclear DNA Amounts in Angiosperms: Progress, Problems and Prospects. Annals of Botany 95: 45-90. https://doi.org/10.1093/aob/mci003
Bennett, M. D. & Leitch, I. J. (2011). Nuclear DNA amounts in angiosperms -targets, trends and tomorrow. Annals of Botany 107: 467-590. https://doi.org/10.1093/aob/mcq258
Bennetzen, J. L. & Kellogg, E. A. (1997). Do plants have a one-way ticket to genomic obesity? Plant Cell 9: 1509-1514. doi: 10.1105/tpc.9.9.1509
Bottini, M. C. J., Greizerstein, E. J., Aulicino, M. B. & Poggio, L. (2000). Relationships among genome size, environmental conditions and geographical distribution in natural populations of NW Patagonian Species of Berberis L. (Berberidaceae). Annals of Botany 86: 565-573. https://doi.org/10.1006/anbo.2000.1218
Brandham, P. E. & Doherty, M. J. (1998). genome size variation in the Aloaceae, an Angiosperm family displaying karyotypic orthoselection. Annals of Botany 82 (A): 67-73. https://doi.org/10.1006/anbo.1998.0742
Bureš, P., Pavlíček, T., Horován, L. & Nevo, E. (2004). Microgeographic genome size differentiation of the carob tree, Ceratonia siliqua, at ‘Evolution Canyon’, Israel. Annals of Botany 93: 529-535. https://doi.org/10.1093/aob/mch074
Bureš, P., Ozcan, M., Ŝmerda, J., Michálková, E., Horová, L., Plačková, K., Ŝmarda, P., Elliott, T. L., Veselý, P., Ćato, S.,… et al. (2023). Evolution of genome size and GC content in the tribe Carduinae (Asteraceae): rare descending dysploidy and polyploidy, limited environmental control and strong phylogenetic signal. Preslia 95: 185-213. https://doi.org/10.23855/preslia.2023.185
Bureš, P., Elliott, T. L., Veselý, P., Ŝmarda, P., Forest, F., Leitch, I. J., Lughadha, E.N., Soto Gomez, M., Pironon, S., Brown, M. J. M., Ŝmerda, J. & Zedek, F. (2024). The global distribution of angiosperm genome size is shaped by climate. New Phytologist 242: 744-759. https://doi.org/10.1111/nph.19544
Burton, T. & Husband, B. (2001). Fecundity and offspring ploidy in matings among diploid, triploid and tetraploid Chamerion angustifolium (Onagraceae): consequences for tetraploid establishment. Heredity 87: 573-582. https://doi.org/10.1046/j.1365-2540.2001.00955.x
Canhos, V. P., Souza, S., & Canhos, D. A. L. (2004). Global biodiversity informatics: Setting the scene for a “new world” of ecological modeling. Biodiversity Informatics 1: 1-13.
Castro, B. M. (2016). Base de datos de biodiversidad de la Comunidad de Madrid y su aplicación a estudios de flora y vegetación. Tesis Doctoral, Universidad Complutense de Madrid, Facultad de Ciencias Biológicas. Madrid, España. https://hdl.handle.net/20.500.14352/26903
Chrtek, Jr. J., Zahradníček, J., Krak, K. & Fehrer, J. (2009). Genome size in Hieracium subgenus Hieracium (Asteraceae) is strongly correlated with major phylogenetic groups. Annals of Botany 104: 161-178. https://doi.org/10.1093/aob/mcp107
De Laat, A. M. A., Göhde, W. & Vogelzang, M. J. D. C. (1987). Determination of ploidy of single plants and plant populations by flow cytometry. Plant Breed 99: 303-307. https://doi.org/10.1111/j.1439-0523.1987.tb01186.x
Dematteis, M. (2009). Revisión taxonómica del género sudamericano Chrysolaena (Vernonieae, Asteraceae). Boletín de la Sociedad Argentina de Botánica 44: 103-170. Disponible en: <https://www.scielo.org.ar/scielo.php?script=sci_arttext&pid=S1851-23722009000100011&lng=es&nrm=iso>. ISSN 1851-2372.
Devos, K., Brown, J. & Bennetzen, J. (2002). Genome size reduction through illegitimate recombination counteracts genome expansion in Arabidopsis. Genome Research 12: 1075-1079. http://www.genome.org/cgi/doi/10.1101/gr.132102
Dimitrova, D. & Greilhuber, J. (2000). Karyotype and DNA content in ten species of Crepis (Asteraceae) distributed in Bulgaria. Journal of Linnean Society 132: 281-297. https://doi.org/10.1111/j.1095-8339.2000.tb01532.x
Doležel, J. & Göhde, W. (1995). Sex determination in dioecious plants Melandrium album and M. rubrum using high‐resolution flow cytometry. Cytometry 19: 103-106. https://doi.org/10.1002/cyto.990190203
Doležel, J., Greilhuber, J., Lucretti, S., Meister, A., Lysák, M. A., Nardi, L. & Obermayer, R. (1998). plant genome size estimation by flow cytometry: Inter-laboratory comparison. Annals of Botany 82: 17-26. https://doi.org/10.1093/oxfordjournals.aob.a010312
Doležel, J., Greilhuber, J. & Suda, J. (2007). Estimation of nuclear DNA content in plants using flow cytometry. Nature Protocols 2: 2233-2244. https://doi.org/10.1038/nprot.2007.310
Dušková, E., Kolář, F., Skelenář, P., Rauchová, J., Kubešová, M., Fér, T., Suda, J. & Marhold, K. (2010). Genome size correlates with growth form, habitat and phylogeny in the Andean genus Lasiocephalus (Asteraceae). Preslia 82: 127-148.
Echeverría, M. L. & Camadro, E. L. (2017). Chromosome numbers, meiotic abnormalities, and 2n pollen formation in accessions of the wild species Chrysolaena flexuosa (Vernonieae, Compositae) from its distribution range in Argentina. Boletín de la Sociedad Argentina de Botánica 52: 737-752. Disponible en: <https://www.scielo.org.ar/scielo.php?script=sci_arttext&pid=S1851-23722017000400010&lng=es&nrm=iso>. ISSN 1851-2372.
Elliott, T. L., Zedek, F., Barrett, B., Bruhl, J., Escudero, M., Hroudová, Z., Joly, S., Larridon, I., Luceño, M., Márquez-Corro, J. I,… et al. (2022). Chromosome size matters: genome evolution in the cyperid clade. Annals of Botany 130: 999-1014. https://doi.org/10.1093/aob/mcac136
Faizullah, L., Morton, J. A., Hersch-Green, E. I., Walczyk, A. M., Leitch, A. R. & Leitch, I. J. (2021). Exploring environmental selection on genome size in angiosperms. Trends in Plant Science 26: 1039-1049. https://doi.org/10.1016/j.tplants.2021.06.001
Flora e Funga do Brasil. Jardim Botânico do Rio de Janeiro. Disponible en: <http://floradobrasil.jbrj.gov.br/ >. Consulta: 27/11/2024
Funk, V. A., Susanna, A., Stuessy, T. F., Bayer, R. J. (eds.). (2009). Systematics, evolution and biogeography of Compositae. Vienna: International Association for Plant Taxonomist.
Galbraith, D. W., Harkis, K. R., Maddox, J. M., Ayres, N. M., Sharma, D. P. & Firoozabady, E. (1983). Rapid flow cytometry analysis of the cell cycle in intact plant tissues. Science 220: 1049-1051. https://doi.org/10.1126/science.220.4601.1049
Garcia, S., Inceer, H., Garnatje, T. & Valles, J. (2005). Genome size variation in some representatives of the genus Tripleurospermum. Biologia Plantarum 49: 381-387. https://doi.org/10.1007/s10535-005-0011-z
Garcia, S., Leitch, I. J., Anadon-Rosell, A., Canela, M. A., Gálvez, F., Garnatje, T., Gras, A., Hidalgo, O., Johnston, E., De Xaxars, G. M., Pellicer, J., Siljak-Yakovlev, S., Vallès, J., Vitales, D. & Bennett, M. D. (2014). Recent updates and developments to plant genome size databases. Nucleic Acids Research 42(D1): 1159-1166. https://doi.org/10.1093/nar/gkt1195
Garnatje, T., Vallès, J., Garcia, S., Hidalgo, O., Sanz, M., Canela, M. A. & Siljak-Yakovlev, S. (2004). Genome size in Echinops L. and related genera (Asteraceae, Cardueae): karyological, ecological and phylogenetic implication. Biology of the Cell 96: 117-124. https://doi.org/10.1016/j.biolcel.2003.11.005
Garnatje, T., Garcia, S., Vilatersana, R. & Vallès, J. (2006). Genome size variation in the genus Carthamus (Asteraceae, Cardueae): Systematic implications and additive changes during allopolyploidization. Annals of Botany 97: 461-467. https://doi.org/10.1093/aob/mcj050
Garnatje, T., Canela, M. A., Garcia, S., Hidalgo, O., Pellicer, J., Sánchez-Jiménez, I., Siljak-Yakovlev, S., Vitales, D. & Vallès, J. (2011). GSAD: a genome size database in the Asteraceae. Cytometry Part A 79A: 401-404. http://doi.org/10.1002/cyto.a.21056
Gasmanová, N., Lebeda, A., Dolezalová, I., Cohen, T., Pavlíček, T., Fahima, T. & Nevo, E. (2007). Genome size variation of Lotus peregrinus at Evolution Canyon I microsite, Lower Nahal Oren, Mt. Carmel, Israel. Acta Biologica Cracoviensia Series Botanica 49: 39-46.
Greilhuber, J. (1998). Intraspecific variation in genome size: a critical reassessment. Annals of Botany 82(A): 27-35. https://doi.org/10.1006/anbo.1998.0725
Greilhuber, J. (2005). Intraspecific variation in genome size in Angiosperms: identifying its existence. Annals of Botany 95: 91-98. https://doi.org/10.1093/aob/mci004
Greilhuber, J. & Leitch, I. J. (2013). Genome size and the phenotype. En Leitch, I. J., Greilhuber, J., Doležel, J. & J. F. Wendel (eds.), Plant Genome Diversity Physical Structure, Behaviour and Evolution of Plant Genomes. Vol. 2, pp. 323-344. Springer Vienna, Austria.
Ingwersen, P. & Chavan, V. (2011). Indicators for the Data Usage Index (DUI): An incentive for publishing primary biodiversity data through global information infrastructure. BMC Bioinformatics (Suppl 15) (S3).
Kalendar, R., Tanskanen, J., Immonen, S., Nevo, E. & Schulman, A. H. (2000). Genome evolution of wild barley (Hordeum spontaneum) by BARE-1 retrotransposon dynamics in response to sharp microclimatic divergence. Proceedings of the National Academy of Sciences USA 97: 6603-6607. https://doi.org/10.1073/pnas.11058749
Keeley, S. C. & Robinson, H. (2009). Vernonieae. En Funk, V. A., Susanna, A., Stuessy, T. F. & R. J. Bayer (eds.), Systematics, evolution and biogeography of Compositae, pp. 439-469: International Association for Plant Taxonomist, Vienna.
Kellogg, E. A. & Bennetzen, J. L. (2004). The evolution of nuclear genome structure in seed plants. American Journal of Botany 91: 1709-1725. https://doi.org/10.3732/ajb.91.10.1709
Knight, C. A. & Ackerly, D. D. (2002). Variation in nuclear DNA content across environmental gradients: a quantile regression analysis. Ecology Letters 5: 66-76. https://doi.org/10.1046/j.1461-0248.2002.00283.x
Leitch, I. J. & Bennett, M. D. (2004). Genome downsizing in polyploid plants. Biological Journal of the Linnean Society 82: 651-663. https://doi.org/10.1111/j.1095-8312.2004.00349.x
Leitch, I. J. & Bennett, M. D. (2007). Genome size and its uses: the impact of flow cytometry. En Doležel, J., Greilhuber, J. & J. Suda (eds.), Flow cytometry with plant cells, pp. 153-176. WileyVCH, Weinheim.
Leitch, A. R. & Leitch, I. J. (2008). Genomic plasticity and the diversity of polyploid plants. Science 320: 481-483. DOI: 10.1126/science.115358
Leitch, I. J. & Leitch, A. R. (2013). Genome size diversity and evolution in land plants. En Greilhuber, J., Doležel, J., Wendel, J. F. & I. J. Leitch (eds.), Plant Genome Diversity Volume 2: Physical Structure, Behaviour and Evolution of Plant Genomes, pp. 307-322. Springer Vienna.
Leitch, I. J., Johnston, E., Pellicer, J., Hidalgo, O., Bennett, M. D. (2019). Angiosperm DNA C-values Database (Release 7.1). https://cvalues.science.kew.org/
Lira-Noriega, A., Soberón, J., Navarro-Sigüenza, A. G.,…et al. (2007). Scale dependency of diversity components estimated from primary biodiversity data and distribution maps. Diversity and Distributions 13: 185-195. https://doi.org/10.1111/j.1472-4642.2006.00304.x
Murray, B. G. (2005). When does intraspecific C-value variation become taxonomically significant? Annals of Botany 95: 119-125. https://doi.org/10.1093/aob/mci007
Naranjo, C. A., Ferrari, M. R., Palermo, A. M. & Poggio, L. (1998). Karyotype, DNA content and meiotic behavior in five South American species of Vicia (Fabaceae). Annals of Botany 82: 757-764. https://doi.org/10.1006/anbo.1998.0744
Oliva Brañas, M. & Vallès, J. (1994). Karyological studies in some taxa of the genus Artemisia L. (Asteraceae). Canadian Journal of Botany 72: 1126-1135. https://doi.org/10.1139/b94-138
Olšavská, K., Perný, M., Španiel, S. & Šingliarová, B. (2012). Nuclear DNA content variation among perennial taxa of the genus Cyanus (Asteraceae) in Central Europe and adjacent areas. Plant Systematics and Evolution 298: 1463–1482. https://doi.org/10.1007/s00606-012-0650-4
Ohri, D. (1998). Genome size variation and plant systematics. Annals of Botany 82: 75-83. https://doi.org/10.1006/anbo.1998.0765
Pavliček, T., Bureš, P., Horová, L., Raskina, O. & Nevo, E. (2008). Genome size microscale divergence of Cyclamen persicum in Evolution Canyon, Israel. Central European Journal of Biology 3: 83-90. https://doi.org/10.2478/s11535-007-0043-9
Pellicer, J., Garcia, S., Garnatje, T., Dariima, S., Korobkov, A. A. & Vallès, J. (2007). Chromosome numbers in some Artemisia (Asteraceae, Anthemideae) species and genome size variation in its subgenus Dracunculus: karyological, systematic and phylogenetic implications. Chromosome Botany 2: 45-53. https://doi.org/10.3199/iscb.2.45
Poggio, L., González, G. & Naranjo, C. A. (2007). Chromosome Studies in Hippeastrum (Amaryllidaceae): variation in genome size. Botanical Journal of Linnean Society 155: 171-178. https://doi.org/10.1111/j.1095-8339.2007.00645.x
Poggio, L., De Sous, M., Batje, N. H., Heuvelink, G. B. M., Kempen, B., Ribeiro, E. & Rossiter, D. (2021). SoilGrids 2.0: producing soil information for the globe with quantified spatial uncertainty. Soil 7: 217-240. https://doi.org/10.5194/soil-7-217-2021
Pringle, G. J. & Murray, B. G. (1993). Karyotypes and C-banding patterns in species of Cyphomandra Mart. ex Sendtner (Solanaceae). Botanical Journal of the Linnean Society 111: 331-342. https://doi.org/10.1111/j.1095-8339.1993.tb01907.x
R CORE TEAM (2024). R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna, Austria. <https://www.R-project.org/>.
Raina, S. N. & Rees, H. (1983). DNA variation between and within chromosome complements of Vicia species. Heredity 51: 335-346. https://doi.org/10.1038/hdy.1983.38
Raina, S. N., Parida, A., Koul, K. K., Salimath, S. S., Bisht, M. S., Raja, V. & Khoshoo, T. N. (1994). Associated chromosomal DNA changes in polyploids. Genome 37: 560-564. https://doi.org/10.1139/g94-080
Rees, H. (1984). Nuclear DNA variation and the homology of chromosomes. En Grant, W. F. (ed.), Plant biosystematics, pp. 87-96. Academic Press, Toronto.
Robinson, H. (1988). Studies in the Lepidaploa complex (Vernonieae: Asteraceae). V. The new genus Chrysolaena. Proceedings of the Biological Society of Washington 100: 952-958.
Robinson, H. (1999). Generic and subtribal classification of American Vernonieae. Smithsonian Contributions of Botany 89: 1-116.
Siljak-Yakovlev, S., Solic, M. E., Catrice, O., Brown, S. C. & Papes, D. (2005). Nuclear DNA content and chromosome number in some diploid and tetraploid Centaurea (Asteraceae: Cardueae) from the Dalmatia Region. Plant Biology 397-404. https://doi.org/10.1055/s-2005-865693
Slovák, M., Vít, P., Urfus, T. & Suda, J. (2009). Complex pattern of genome size variation in a polymorphic member of the Asteraceae. Journal of Biogeography 36: 372-384. https://doi.org/10.1111/j.1365-2699.2008.02005.x
Šmarda, P. & Bureš, P. (2006). Intraspecific DNA content variability in Festuca pallens on different geographical scales and ploidy levels. Annals of Botany 98: 665-678. https://doi.org/10.1093/aob/mcl150
Šmarda, P., Bureš, P. & Hovrová, L. (2007). Random distribution pattern and non-adaptivity of genome size in a highly variable population of Festuca pallens. Annals of Botany 100: 141-150. https://doi.org/10.1093/aob/mcm095
Šmarda, P., Horová, L., Bureš, P., Hralová, I. & Marková, M. (2010). Stabilizing selection on genome size in a population of Festuca pallens under conditions of intensive intraspecific competition. New Phytologist 187: 1195-1204. https://doi.org/10.1111/j.1469-8137.2010.03335.x
Šmarda, P., Knápek, O., Březinová, A., Horová, L., Grulich, V., Danihelka, J., Veselý, P., Šmerda, J., Rotreklová, O. & Bureš, P. (2019). Genome sizes and genomic guanine+cytosine (GC) contents of the Czech vascular flora with new estimates for 1700 species. Preslia 91: 117-142. https://doi.org/10.23855/preslia.2019.117
Soares, P. N., Gonçalves-Esteves, V., Semir, J. & Nakajima, J. N. (2020). Chrysolaena glandulosa (Vernonieae, Asteraceae): a new species from Brazil. Phytotaxa 439: 295-300. https://doi.org/10.11646/phytotaxa.439.3.11
Suda, J., Krahulcová, A., Trávníček, P., Rosenbaumová, R., Peckert, T., Krahulec, F. (2007). Genome size variation and species relationships in Hieracium sub-genus Pilosella (Asteraceae) as inferred by flow cytometry. Annals of Botany 100: 1323-1335. https://doi.org/10.1093/aob/mcm218
Torrell, M., Bosch, M., Martín, J. & Vallès, J. (1999). Cytogenetic and isozymic characterization of the narrow endemic species Artemisia molinieri Quézel, Barbero & R. Loisel (Asteraceae, Anthemideae); implications for its systematics and conservation. Canadian Journal of Botany 77: 1-10. https://doi.org/10.1139/b98-193
Torrell, M. & Vallès, J. (2001). Genome size in 21 Artemisia L. species (Asteraceae, Anthemideae): Systematic, evolutionary and ecological implications. Genome 44: 231- 238. https://doi.org/10.1139/g01-004
Valdecasas, A. G. & Camacho, A. (2003). Conservation to the rescue of taxonomy. Biodiversity and Conservation 12: 1113-1117. https://doi.org/10.1023/A:1023082606162
Vega, A. J. & Dematteis, M. (2016). Cytotaxonomy of some species of Vernonanthura and Vernonia (Asteraceae, Vernonieae) from South America. Caryologia 69: 29-37. https://doi.org/10.1080/00087114.2015.1109937
Vega, G. C., Pertierra, L. R. & Olalla-Tárraga, M. A. (2018). Data from: MERRAclim, a high-resolution global dataset of remotely sensed bioclimatic variables for ecological modelling [Dataset]. Dryad. https://doi.org/10.5061/dryad.s2v81
Via Do Pico, G. M. (2015) Estudios biosistemáticos en especies del género Chrysolaena (Vernonieae, Asteraceae). Tesis Doctoral, Universidad Nacional de Córdoba, Argentina. 285 pp.
Via Do Pico, G. M. & Dematteis, M. (2012). Chromosome number, meiotic behavior and pollen fertility of six species of Chrysolaena (Vernonieae, Asteraceae). Caryologia 65: 176-181. https://doi.org/10.1080/00087114.2012.726506
Via Do Pico, G. M. & Dematteis, M. (2013). Karyotype analysis and DNA content in some species of Chrysolaena (Vernonieae, Asteraceae). Plant Biosystems 147: 864-873. https://doi.org/10.1080/11263504.2012.751068
Via Do Pico, G. M. & Dematteis, M. (2014). Cytotaxonomy of two species of genus Chrysolaena H. Robinson, 1988 (Vernonieae, Asteraceae) from Northeast Paraguay. Comparative Cytogenetics 8: 125-137. https://doi.org/10.3897/CompCytogen.v8i2.7209
Via Do Pico, G. M. & Dematteis, M. (2017). Meiotic behaviour and B chromosomes in Argentine populations of Chrysolaena verbascifolia (Vernonieae: Asteraceae). Webbia 72: 139-147. https://doi.org/10.1080/00837792.2016.1260881
Via Do Pico, G. M., Pérez, Y. J., Angulo, M. B. & Dematteis, M. (2019). Cytotaxonomy and geographic distribution of cytotypes of species of the South American genus Chrysolaena (Vernonieae, Asteraceae). Journal of Systematics and Evolution 57: 451-467. https://doi.org/10.1111/jse.12471
Walker, D. J. Moñino, I. & Correal, E. (2006). Genome size in Bituminaria bituminosa (L.) C.H. Stirton (Fabaceae) populations: separation of “true” differences from environmental effects on DNA determination. Environmental and Experimental Botany 55: 258-265. https://doi.org/10.1016/j.envexpbot.2004.11.005
Wickham, H. (2016). ggplot2: Elegant Graphics for Data Analysis. Springer-Verlag New York. ISBN 978-3-319-24277-4, https://ggplot2.tidyverse.org.).
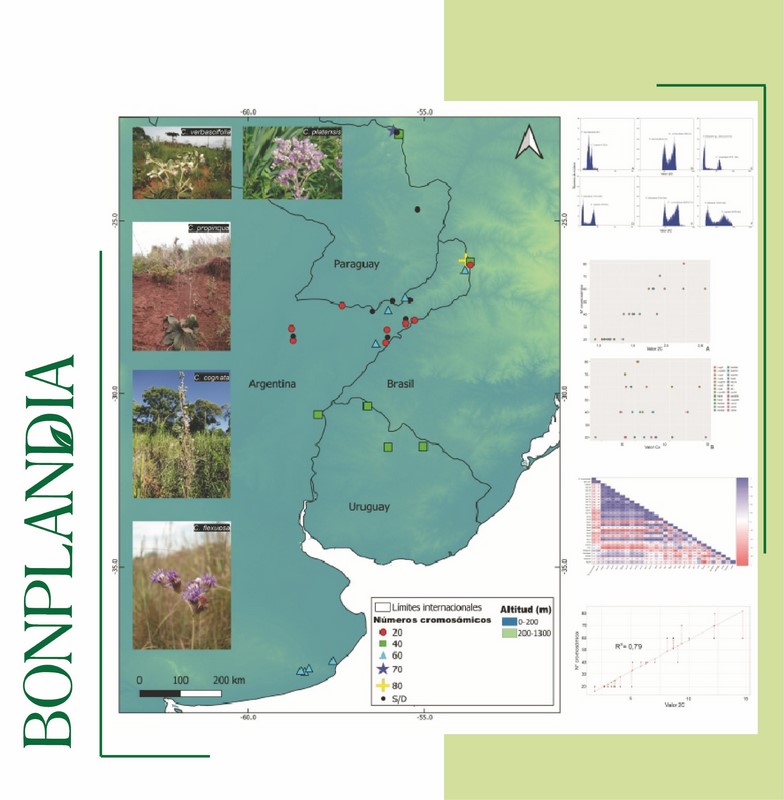
Downloads
Published
How to Cite
Issue
Section
License
Copyright (c) 2026 Bonplandia

This work is licensed under a Creative Commons Attribution 4.0 International License.
Declaration of Adhesion to Open Access
- All contents of Bonplandia journal are available online, open to all and for free, before they are printed.
Copyright Notice
- Bonplandia magazine allows authors to retain their copyright without restrictions.
- The journal is under a Creative Commons Attribution 4.0 International license.














.jpg)


